research interests
To best describe my academic research, I should probably say that I am a “computational material scientist” or a “mechano-chemist”. I am interested in exploring and explaining how properties emerge in matter across scales, mostly using computer simulation. Through different encouters and short-term research positions, I ended up studying a wide spectrum of materials and systems, from stability of concrete structures under seismic loading, to migration of mesenchymal stem cells, and currently, failure of polymer nanocomposites. I am now getting more and more interested in studying biological materials, in particular those with outstanding mechanical properties, which all started by observing structure-properties relationships in the material of choice in the animal kingdom: collagen.
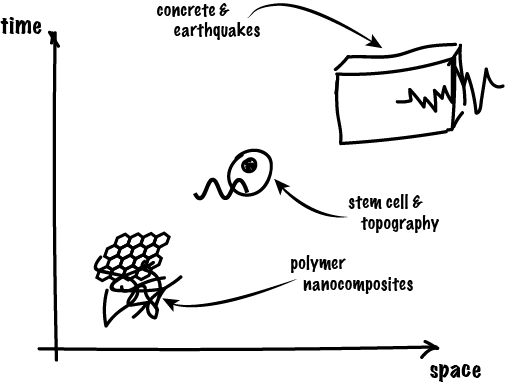 hierarchies of my research projects: from molecular networks of epoxy and graphene sheets to massive concrete structures, passing by microscopic human cells.
hierarchies of my research projects: from molecular networks of epoxy and graphene sheets to massive concrete structures, passing by microscopic human cells.
Building on the experience of these different projects, I now aim at pursuing my research in the field of biophysics, and in particular mechanobiology. I want to understand how cells and their environment interact, with an emphasis on mechanical cues. My approach resides in using computational models to formalise and test theories or hypotheses drawn from experimental observations. As a principle the complex cell behaviour, during mitosis, migration and so on, can be pictured as combination of rather simple physical mechanisms, aimed at optimising a few biological patterns.